SecStrAnnotator is a software tool for annotation (naming) of secondary structure elements (SSEs), namely helices and β-strands, in protein structures. You can either download the desktop version of SecStrAnnotator, or use SecStrAnnotator Online on this site.
The SSE annotation is template-based, meaning that a structurally similar template protein with annotated SSEs must be provided. SecStrAnnotator Online allows automatic annotation using our pre-defined template for cytochromes P450 (CYPs) or by user-defined template.
With SecStrAPI plugin you can view precomputed CYP annotation directly in PyMOL.
Annotate
Annotate your own structures with SecStrAnnotator ⇒
Examples
Bacterial CYP
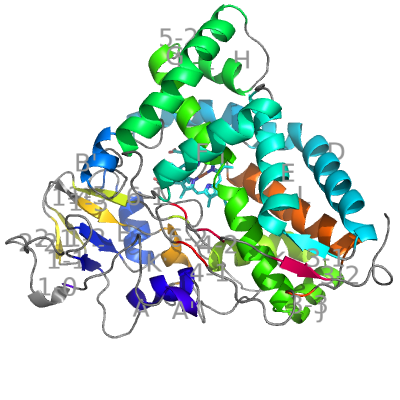
Cytochrome P450cam from Pseudomonas putida was the first CYP with experimentally determined structure. Hence it served as the base for the SSE nomenclature: 12 major helices were named A–L and the β-sheets were numbered 1, 2, 3... Over the years, shorter helices (A′, B′ B″...) have been added to this nomenclature convention.
Eukaryotic CYP
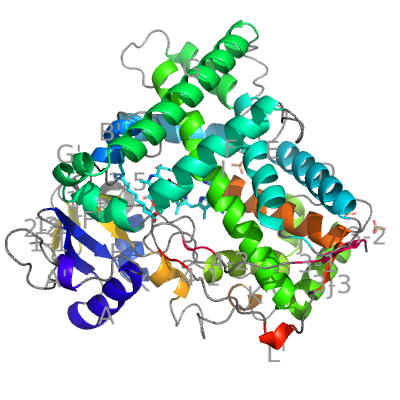
The eukaryotic CYPs differ from the bacterial in several regions of their structure. The most obvious are two extra helices F′ and G′ between helices F and G, and longer helix J (15 residues) followed an extra helix J′. As an example of eukaryotic CYPs we show CYP2C8 from Homo sapiens (which also serves as the annotation template for other CYPs).
Anomalous bacterial CYP
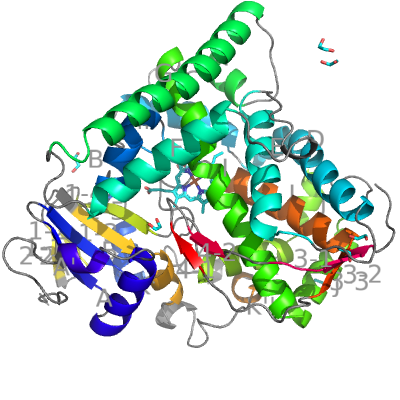
CYP BM3 from Bacillus megaterium is one of the few known bacterial CYPs, which show some typically eukaryotic structural features. Like eukaryotic CYPs, it has a long helix J (15 residues) and helix J′. Like bacterial CYPs, it contains no F′ and G′ helices. CYP BM3 is also interesting because besides the CYP domain it contains a NADPH-dependent cytochrome P450 reductase domain (not covered in this example).
Citing
If you found SecStrAnnotator helpful, please cite:
- Midlik A, Navrátilová V, Moturu TR, Koča J, Svobodová R, Berka K (2021) Uncovering of cytochrome P450 anatomy by SecStrAnnotator. Sci Rep, 11, 12345. https://doi.org/10.1038/s41598-021-91494-8
- Midlik A, Hutařová Vařeková I, Hutař J, Moturu TR, Navrátilová V, Koča J, Berka K, Svobodová Vařeková R (2019) Automated family-wide annotation of secondary structure elements. In: Kister AE (ed) Protein supersecondary structures: Methods and protocols, Humana Press. ISBN 978-1-4939-9160-0. https://doi.org/10.1007/978-1-4939-9161-7_3
Contact
If you have any feedback to SecStrAnnotator or suggestions for protein families for annotation, please contact us:
Adam Midlik, midlik@mail.muni.cz, Structural bioinformatics research group, Masaryk University